Interval-Wise Testing for Omics Data
Preface
IWTomics (Cremona et al. 2018) implements the Interval-Wise Testing (IWT; Pini and Vantini 2017) for omics data. The protocol infers differences in the "omics" data between two-sets of genomic regions and doesn't even require manipulating them for comparative scale.
The data used in this exercise is of endogeneous retroviruses in mouse. The agenda is to compare the recombination hotspots in the flanking regions of fixed ETn versus the control regions. Apart from the two region-sets, the dataset also contains a feature “Recombination hotspots content”. More details about the dataset can be viewed in the formal tutorial [1].
Tools In-Use
The following tools are needed to be installed.
- IWTomics Load tool from the iwtomics_loadandplot repository.
Loading Data
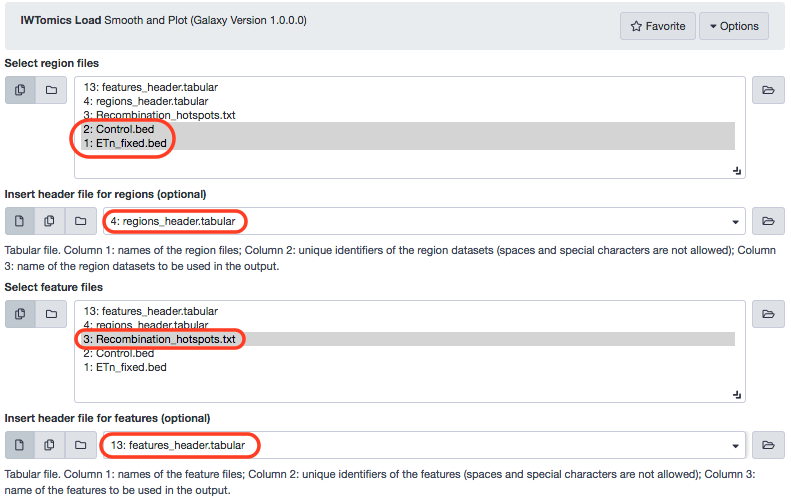
Let us commence by loading data into a new session. There are two region-sets, a feature file, and two header files, in that order.
- Fixed ETn regions https://zenodo.org/record/1288429/files/ETn_fixed.bed
- Control regions https://zenodo.org/record/1288429/files/Control.bed
- Recombination Hotspots https://zenodo.org/record/1288429/files/Recombination_hotspots.txt
- Regions' header https://zenodo.org/record/1288429/files/regions_header.tabular
- Features' header https://zenodo.org/record/1288429/files/features_header.tabular
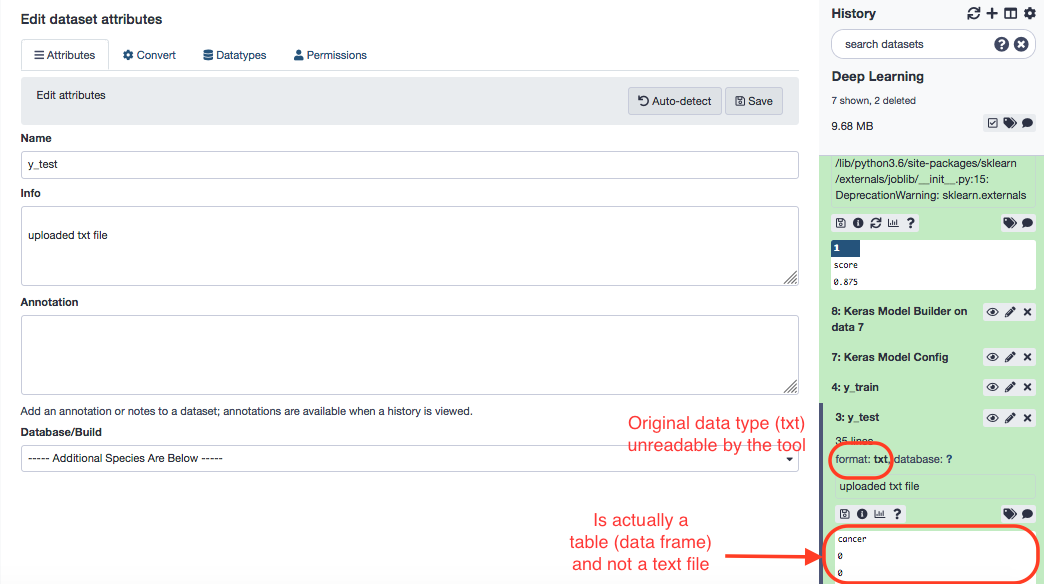
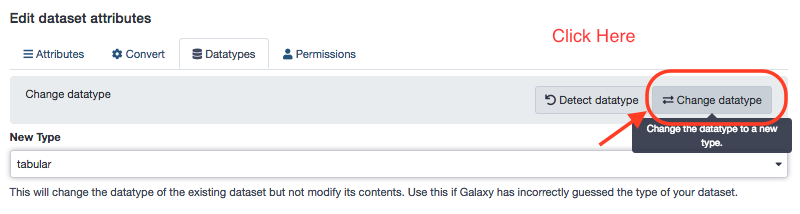
P.S. The "Features' header" file is a txt file and might not be workable. We shall choose to edit attribues for that file. It is a general scenario in data handling, as most of the data is in the form of tables (tabular) and if supplied as a text file, is not interpreted appropriately by the method. To change the txt file to a tabular format, Galaxy provides for a built-in, data-type conversion facility; one can choose the "Edit attributes" (Pencil icon) for the file and proceed as follows. Also, this example is not the part of this workflow ; though the file features_header.tabular can be dealt in a similar fashion after downloading from the source.
Look for the new data-type. All major data formats are supported by Galaxy.
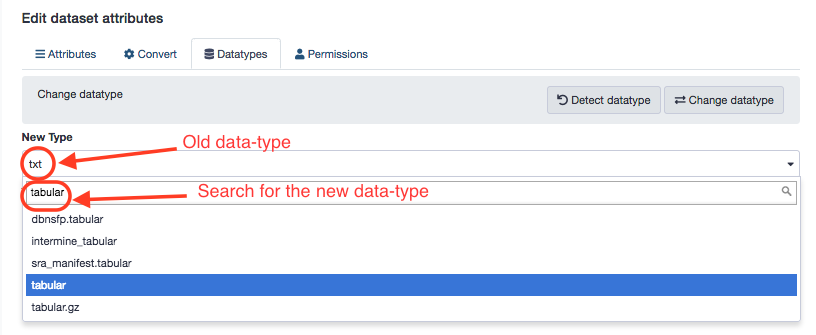
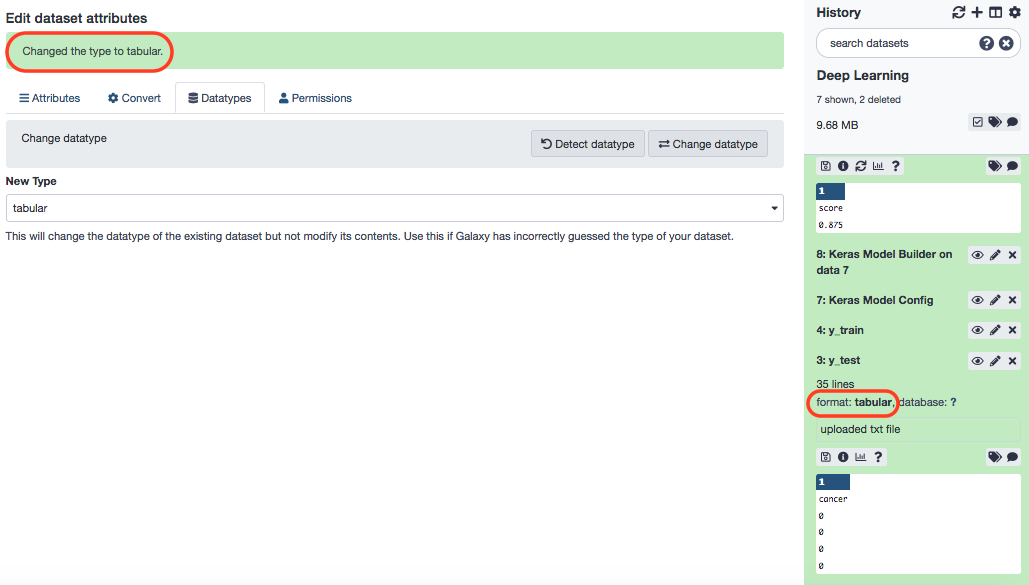
If everything goes well, you have the original file with a changed profile.
With the file attribute changed, we can now move further with executing the tool. The remaining options set to default, we shall indicate the following input paramaters.
References
- Marzia A Cremona, Fabio Cumbo, 2020 Interval-Wise Testing for omics data (Galaxy Training Materials). /training-material/topics/statistics/tutorials/iwtomics/tutorial.html Online; accessed Mon Jul 27 2020